Bonus Tutorial: Spike-timing dependent plasticity (STDP)#
Week 2, Day 3: Biological Neuron Models
By Neuromatch Academy
Content creators: Qinglong Gu, Songtin Li, John Murray, Richard Naud, Arvind Kumar
Content reviewers: Spiros Chavlis, Lorenzo Fontolan, Richard Gao, Matthew Krause
Production editors: Gagana B, Spiros Chavlis
Tutorial Objectives#
In this tutorial, we will focus on building a model of a synapse in which its synaptic strength changes as a function of the relative timing (i.e., time difference) between the spikes of the presynaptic and postsynaptic neurons, respectively. This change in the synaptic weight is known as spike-timing dependent plasticity (STDP).
Our goals for this tutorial are to:
build a model of synapse that show STDP
study how correlations in input spike trains influence the distribution of synaptic weights
Towards these goals, we will model the presynaptic input as Poisson type spike trains. The postsynaptic neuron will be modeled as an LIF neuron (see Tutorial 1).
Throughout this tutorial, we assume that a single postsynaptic neuron is driven by \(N\) presynaptic neurons. That is, there are \(N\) synapses, and we will study how their weights depend on the statistics or the input spike trains and their timing with respect to the spikes of the postsynaptic neuron.
Setup#
Install and import feedback gadget#
Show code cell source
# @title Install and import feedback gadget
!pip3 install vibecheck datatops --quiet
from vibecheck import DatatopsContentReviewContainer
def content_review(notebook_section: str):
return DatatopsContentReviewContainer(
"", # No text prompt
notebook_section,
{
"url": "https://pmyvdlilci.execute-api.us-east-1.amazonaws.com/klab",
"name": "neuromatch_cn",
"user_key": "y1x3mpx5",
},
).render()
feedback_prefix = "W2D3_T4_Bonus"
# Import libraries
import matplotlib.pyplot as plt
import numpy as np
import time
Figure Settings#
Show code cell source
# @title Figure Settings
import logging
logging.getLogger('matplotlib.font_manager').disabled = True
import ipywidgets as widgets # interactive display
%config InlineBackend.figure_format='retina'
# use NMA plot style
plt.style.use("https://raw.githubusercontent.com/NeuromatchAcademy/course-content/main/nma.mplstyle")
my_layout = widgets.Layout()
Plotting functions#
Show code cell source
# @title Plotting functions
def my_raster_plot(range_t, spike_train, n):
"""Generates poisson trains
Args:
range_t : time sequence
spike_train : binary spike trains, with shape (N, Lt)
n : number of Poisson trains plot
Returns:
Raster_plot of the spike train
"""
# Find the number of all the spike trains
N = spike_train.shape[0]
# n should be smaller than N:
if n > N:
print('The number n exceeds the size of spike trains')
print('The number n is set to be the size of spike trains')
n = N
# Raster plot
i = 0
while i <= n:
if spike_train[i, :].sum() > 0.:
t_sp = range_t[spike_train[i, :] > 0.5] # spike times
plt.plot(t_sp, i * np.ones(len(t_sp)), 'k|', ms=10, markeredgewidth=2)
i += 1
plt.xlim([range_t[0], range_t[-1]])
plt.ylim([-0.5, n + 0.5])
plt.xlabel('Time (ms)')
plt.ylabel('Neuron ID')
plt.show()
def my_example_P(pre_spike_train_ex, pars, P):
"""Generates two plots (raster plot and LTP vs time plot)
Args:
pre_spike_train_ex : spike-train
pars : dictionary with the parameters
P : LTP ratio
Returns:
my_example_P returns a rastert plot (top),
and a LTP ratio across time (bottom)
"""
spT = pre_spike_train_ex
plt.figure(figsize=(7, 6))
plt.subplot(211)
color_set = ['red', 'blue', 'black', 'orange', 'cyan']
for i in range(spT.shape[0]):
t_sp = pars['range_t'][spT[i, :] > 0.5] # spike times
plt.plot(t_sp, i*np.ones(len(t_sp)), '|',
color=color_set[i],
ms=10, markeredgewidth=2)
plt.xlabel('Time (ms)')
plt.ylabel('Neuron ID')
plt.xlim(0, 200)
plt.subplot(212)
for k in range(5):
plt.plot(pars['range_t'], P[k, :], color=color_set[k], lw=1.5)
plt.xlabel('Time (ms)')
plt.ylabel('P(t)')
plt.xlim(0, 200)
plt.tight_layout()
plt.show()
def mySTDP_plot(A_plus, A_minus, tau_stdp, time_diff, dW):
plt.figure()
plt.plot([-5 * tau_stdp, 5 * tau_stdp], [0, 0], 'k', linestyle=':')
plt.plot([0, 0], [-A_minus, A_plus], 'k', linestyle=':')
plt.plot(time_diff[time_diff <= 0], dW[time_diff <= 0], 'ro')
plt.plot(time_diff[time_diff > 0], dW[time_diff > 0], 'bo')
plt.xlabel(r't$_{\mathrm{pre}}$ - t$_{\mathrm{post}}$ (ms)')
plt.ylabel(r'$\Delta$W', fontsize=12)
plt.title('Biphasic STDP', fontsize=12, fontweight='bold')
plt.show()
Helper functions#
Show code cell source
# @title Helper functions
def default_pars_STDP(**kwargs):
pars = {}
# typical neuron parameters
pars['V_th'] = -55. # spike threshold [mV]
pars['V_reset'] = -75. # reset potential [mV]
pars['tau_m'] = 10. # membrane time constant [ms]
pars['V_init'] = -65. # initial potential [mV]
pars['V_L'] = -75. # leak reversal potential [mV]
pars['tref'] = 2. # refractory time (ms)
# STDP parameters
pars['A_plus'] = 0.008 # magnitude of LTP
pars['A_minus'] = pars['A_plus'] * 1.10 # magnitude of LTD
pars['tau_stdp'] = 20. # STDP time constant [ms]
# simulation parameters
pars['T'] = 400. # Total duration of simulation [ms]
pars['dt'] = .1 # Simulation time step [ms]
# external parameters if any
for k in kwargs:
pars[k] = kwargs[k]
pars['range_t'] = np.arange(0, pars['T'], pars['dt']) # Vector of discretized time points [ms]
return pars
def Poisson_generator(pars, rate, n, myseed=False):
"""Generates poisson trains
Args:
pars : parameter dictionary
rate : noise amplitute [Hz]
n : number of Poisson trains
myseed : random seed. int or boolean
Returns:
pre_spike_train : spike train matrix, ith row represents whether
there is a spike in ith spike train over time
(1 if spike, 0 otherwise)
"""
# Retrieve simulation parameters
dt, range_t = pars['dt'], pars['range_t']
Lt = range_t.size
# set random seed
if myseed:
np.random.seed(seed=myseed)
else:
np.random.seed()
# generate uniformly distributed random variables
u_rand = np.random.rand(n, Lt)
# generate Poisson train
poisson_train = 1. * (u_rand < rate * (dt / 1000.))
return poisson_train
Section 1: Spike-timing dependent plasticity (STDP)#
Video 1: STDP#
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_STDP_Video")
Model of STDP#
The phenomenology of STDP is generally described as a biphasic exponentially decaying function. That is, the instantaneous change in weights is given by:
where \(\Delta W\) denotes the change in the synaptic weight, \(A_+\) and \(A_-\) determine the maximum amount of synaptic modification (which occurs when the timing difference between presynaptic and postsynaptic spikes is close to zero), \(\tau_+\) and \(\tau_-\) determine the ranges of pre-to-postsynaptic interspike intervals over which synaptic strengthening or weakening occurs. Thus, \(\Delta W > 0 \) means that postsynaptic neuron spikes after the presynaptic neuron.
This model captures the phenomena that repeated occurrences of presynaptic spikes within a few milliseconds before postsynaptic action potentials lead to long-term potentiation (LTP) of the synapse, whereas repeated occurrences of presynaptic spikes after the postsynaptic ones lead to long-term depression (LTD) of the same synapse.
The latency between presynaptic and postsynaptic spike (\(\Delta t\)) is defined as:
where \(t_{\rm pre}\) and \(t_{\rm post}\) are the timings of the presynaptic and postsynaptic spikes, respectively.
Complete the following code to set the STDP parameters and plot the STDP function. Note that for simplicity, we assume \(\tau_{+} = \tau_{-} = \tau_{\rm stdp}\).
Coding Exercise 1A: Compute the STDP changes \(\Delta W\)#
Note, as shown above, the expression of \(\Delta W\) is different for \(t_{post}>t_{pre}\) and \(t_{post}<t_{pre}\). In the code, we use the parameter time_diff that describes the \(t_{pre}-t_{post}\), as given above.
After implementing the code, you can visualize the STDP kernel, which describes how much the synaptic weight will change given a latency between the presynaptic and postsynaptic spikes.
def Delta_W(pars, A_plus, A_minus, tau_stdp):
"""
Plot STDP biphasic exponential decaying function
Args:
pars : parameter dictionary
A_plus : (float) maximum amount of synaptic modification
which occurs when the timing difference between pre- and
post-synaptic spikes is positive
A_minus : (float) maximum amount of synaptic modification
which occurs when the timing difference between pre- and
post-synaptic spikes is negative
tau_stdp : the ranges of pre-to-postsynaptic interspike intervals
over which synaptic strengthening or weakening occurs
Returns:
dW : instantaneous change in weights
"""
#######################################################################
## TODO for students: compute dP, then remove the NotImplementedError #
# Fill out when you finish the function
raise NotImplementedError("Student exercise: compute dW, the change in weights!")
#######################################################################
# STDP change
dW = np.zeros(len(time_diff))
# Calculate dW for LTP
dW[time_diff <= 0] = ...
# Calculate dW for LTD
dW[time_diff > 0] = ...
return dW
pars = default_pars_STDP()
# Get parameters
A_plus, A_minus, tau_stdp = pars['A_plus'], pars['A_minus'], pars['tau_stdp']
# pre_spike time - post_spike time
time_diff = np.linspace(-5 * tau_stdp, 5 * tau_stdp, 50)
dW = Delta_W(pars, A_plus, A_minus, tau_stdp)
mySTDP_plot(A_plus, A_minus, tau_stdp, time_diff, dW)
Example output:
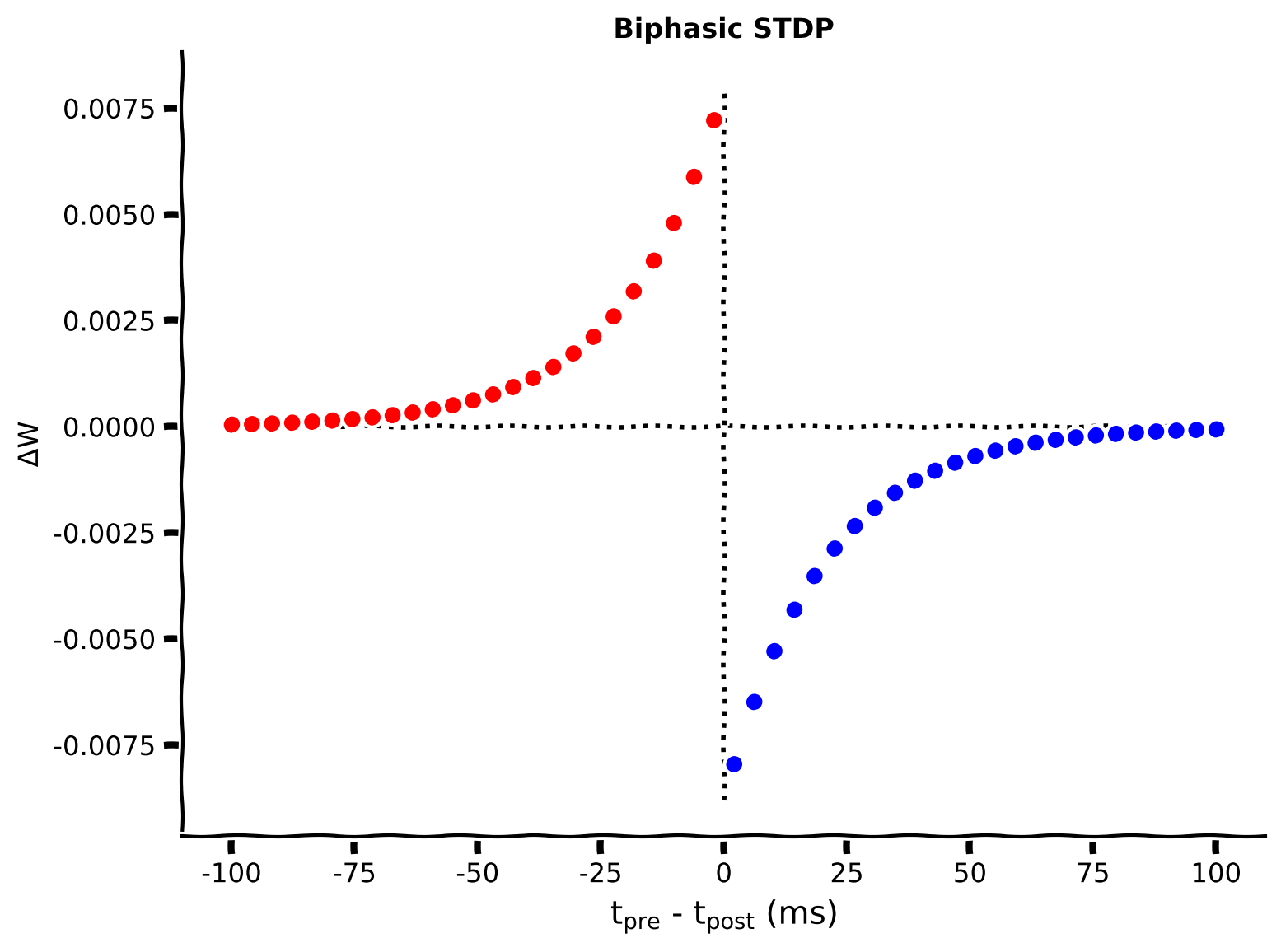
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Compute_STDP_changes_Exercise")
Keeping track of pre- and postsynaptic spikes#
Since a neuron will receive numerous presynaptic spike inputs, in order to implement STDP by taking into account different synapses, we first have to keep track of the pre- and postsynaptic spike times throughout the simulation.
A convenient way to do this is to define the following equation for each postsynaptic neuron:
and whenever the postsynaptic neuron spikes,
This way \(M(t)\) tracks the number of postsynaptic spikes over the timescale \(\tau_{-}\).
Similarly, for each presynaptic neuron, we define:
and whenever there is spike on the presynaptic neuron,
The variables \(M(t)\) and \(P(t)\) are very similar to the equations for the synaptic conductances, i.e., \(g_{i}(t)\), except that they are used to keep track of pre- and postsynaptic spike times on a much longer timescale. Note that, \(M(t)\) is always negative, and \(P(t)\) is always positive. You can probably already guess that \(M\) is used to induce LTD and \(P\) to induce LTP because they are updated by \(A_{-}\) and \(A_{+}\), respectively.
Important note: \(P(t)\) depends on the presynaptic spike times. If we know the presynaptic spike times, \(P\) can be generated before simulating the postsynaptic neuron and the corresponding STDP weights.
Visualization of \(P\)#
Here, we will consider a scenario in which there is a single postsynaptic neuron connected to \(N\) presynaptic neurons.
For instance, we have one postsynaptic neuron which receives Poisson type spiking inputs from five presynaptic neurons.
We can simulate \(P\) for each one of the presynaptic neurons.
Coding Exercise 1B: Compute \(dP\)#
Here, yet again, we use the Euler scheme, which has been introduced several times in the previous tutorials.
Similar to the dynamics of the membrane potential in the LIF model, in a time step \(dt\), \(P(t)\) will decrease by an amount of \(\displaystyle{\frac{dt}{\tau_+}P(t)}\). Whereas, if a presynaptic spike arrives, \(P(t)\) will instantaneously increase by an amount of \(A_+\). Therefore,
where the \(\text{sp_or_not}\) is a binary variable, i.e., \(\text{sp_or_not}=1\) if there is a spike within \(dt\), and \(\text{sp_or_not}=0\) otherwise.
def generate_P(pars, pre_spike_train_ex):
"""
track of pre-synaptic spikes
Args:
pars : parameter dictionary
pre_spike_train_ex : binary spike train input from
presynaptic excitatory neuron
Returns:
P : LTP ratio
"""
# Get parameters
A_plus, tau_stdp = pars['A_plus'], pars['tau_stdp']
dt, range_t = pars['dt'], pars['range_t']
Lt = range_t.size
# Initialize
P = np.zeros(pre_spike_train_ex.shape)
for it in range(Lt - 1):
#######################################################################
## TODO for students: compute dP, then remove the NotImplementedError #
# Fill out when you finish the function
raise NotImplementedError("Student exercise: compute P, the change of presynaptic spike")
#######################################################################
# Calculate the delta increment dP
dP = ...
# Update P
P[:, it + 1] = P[:, it] + dP
return P
pars = default_pars_STDP(T=200., dt=1.)
pre_spike_train_ex = Poisson_generator(pars, rate=10, n=5, myseed=2020)
P = generate_P(pars, pre_spike_train_ex)
my_example_P(pre_spike_train_ex, pars, P)
Example output:

Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Compute_dP_Exercise")
Section 2: Implementation of STDP#
Finally, to implement STDP in spiking networks, we will change the value of the peak synaptic conductance based on the presynaptic and postsynaptic timing, thus using the variables \(P(t)\) and \(M(t)\).
Each synapse \(i\) has its own peak synaptic conductance (\(\bar g_i\)), which may vary between \([0, \bar g_{max}]\), and will be modified depending on the presynaptic and postsynaptic timing.
When the \(i_{th}\) presynaptic neuron elicits a spike, its corresponding peak conductance is updated according to the following equation:
(305)#\[\begin{equation} \bar g_i = \bar g_i + M(t)\bar g_{max} \end{equation}\]Note that, \(M(t)\) tracks the time since the last postsynaptic potential and is always negative. Hence, if the postsynaptic neuron spikes shortly before the presynaptic neuron, the above equation shows that the peak conductance will decrease.
When the postsynaptic neuron spikes, the peak conductance of each synapse is updated according to:
(306)#\[\begin{equation} \bar g_i = \bar g_i + P_i(t)\bar g_{max}, \forall i \end{equation}\]Note that, \(P_i(t)\) tracks the time since the last spike of \(i_{th}\) pre-synaptic neuron and is always positive.
Thus, the equation given above shows that if the presynaptic neuron spikes before the postsynaptic neuron, its peak conductance will increase.
LIF neuron connected with synapses that show STDP#
In the following exercise, we connect \(N\) presynaptic neurons to a single postsynaptic neuron. We do not need to simulate the dynamics of each presynaptic neuron as we are only concerned about their spike times. So, we will generate \(N\) Poisson type spikes. Here, we will assume that all these inputs are excitatory.
We need to simulate the dynamics of the postsynaptic neuron as we do not know its spike times. We model the postsynaptic neuron as an LIF neuron receiving only excitatory inputs.
where the total excitatory synaptic conductance \(g_{E}(t)\) is given by:
While simulating STDP, it is important to make sure that \(\bar g_i\) never goes outside of its bounds.
Function for LIF neuron with STDP synapses#
Show code cell source
# @title Function for LIF neuron with STDP synapses
def run_LIF_cond_STDP(pars, pre_spike_train_ex):
"""
conductance-based LIF dynamics
Args:
pars : parameter dictionary
pre_spike_train_ex : spike train input from presynaptic excitatory neuron
Returns:
rec_spikes : spike times
rec_v : mebrane potential
gE : postsynaptic excitatory conductance
"""
# Retrieve parameters
V_th, V_reset = pars['V_th'], pars['V_reset']
tau_m = pars['tau_m']
V_init, V_L = pars['V_init'], pars['V_L']
gE_bar, VE, tau_syn_E = pars['gE_bar'], pars['VE'], pars['tau_syn_E']
gE_init = pars['gE_init']
tref = pars['tref']
A_minus, tau_stdp = pars['A_minus'], pars['tau_stdp']
dt, range_t = pars['dt'], pars['range_t']
Lt = range_t.size
P = generate_P(pars, pre_spike_train_ex)
# Initialize
tr = 0.
v = np.zeros(Lt)
v[0] = V_init
M = np.zeros(Lt)
gE = np.zeros(Lt)
gE_bar_update = np.zeros(pre_spike_train_ex.shape)
gE_bar_update[:, 0] = gE_init # note: gE_bar is the maximum value
# simulation
rec_spikes = [] # recording spike times
for it in range(Lt - 1):
if tr > 0:
v[it] = V_reset
tr = tr - 1
elif v[it] >= V_th: # reset voltage and record spike event
rec_spikes.append(it)
v[it] = V_reset
M[it] = M[it] - A_minus
gE_bar_update[:, it] = gE_bar_update[:, it] + P[:, it] * gE_bar
id_temp = gE_bar_update[:, it] > gE_bar
gE_bar_update[id_temp, it] = gE_bar
tr = tref / dt
# update the synaptic conductance
M[it + 1] = M[it] - dt / tau_stdp * M[it]
gE[it + 1] = gE[it] - (dt / tau_syn_E) * gE[it] + (gE_bar_update[:, it] * pre_spike_train_ex[:, it]).sum()
gE_bar_update[:, it + 1] = gE_bar_update[:, it] + M[it]*pre_spike_train_ex[:, it]*gE_bar
id_temp = gE_bar_update[:, it + 1] < 0
gE_bar_update[id_temp, it + 1] = 0.
# calculate the increment of the membrane potential
dv = (-(v[it] - V_L) - gE[it + 1] * (v[it] - VE)) * (dt / tau_m)
# update membrane potential
v[it + 1] = v[it] + dv
rec_spikes = np.array(rec_spikes) * dt
return v, rec_spikes, gE, P, M, gE_bar_update
print(help(run_LIF_cond_STDP))
Help on function run_LIF_cond_STDP in module __main__:
run_LIF_cond_STDP(pars, pre_spike_train_ex)
conductance-based LIF dynamics
Args:
pars : parameter dictionary
pre_spike_train_ex : spike train input from presynaptic excitatory neuron
Returns:
rec_spikes : spike times
rec_v : mebrane potential
gE : postsynaptic excitatory conductance
None
Evolution of excitatory synaptic conductance#
In the following, we will simulate an LIF neuron receiving input from \(N=300\) presynaptic neurons.
pars = default_pars_STDP(T=200., dt=1.) # Simulation duration 200 ms
pars['gE_bar'] = 0.024 # max synaptic conductance
pars['gE_init'] = 0.024 # initial synaptic conductance
pars['VE'] = 0. # [mV] Synapse reversal potential
pars['tau_syn_E'] = 5. # [ms] EPSP time constant
# generate Poisson type spike trains
pre_spike_train_ex = Poisson_generator(pars, rate=10, n=300, myseed=2020)
# simulate the LIF neuron and record the synaptic conductance
v, rec_spikes, gE, P, M, gE_bar_update = run_LIF_cond_STDP(pars,
pre_spike_train_ex)
Figures of the evolution of synaptic conductance#
Run this cell to see the figures!
Show code cell source
# @title Figures of the evolution of synaptic conductance
# @markdown Run this cell to see the figures!
plt.figure(figsize=(12., 8))
plt.subplot(321)
dt, range_t = pars['dt'], pars['range_t']
if rec_spikes.size:
sp_num = (rec_spikes / dt).astype(int) - 1
v[sp_num] += 10 # add artificial spikes
plt.plot(pars['range_t'], v, 'k')
plt.xlabel('Time (ms)')
plt.ylabel('V (mV)')
plt.subplot(322)
# Plot the sample presynaptic spike trains
my_raster_plot(pars['range_t'], pre_spike_train_ex, 10)
plt.subplot(323)
plt.plot(pars['range_t'], M, 'k')
plt.xlabel('Time (ms)')
plt.ylabel('M')
plt.subplot(324)
for i in range(10):
plt.plot(pars['range_t'], P[i, :])
plt.xlabel('Time (ms)')
plt.ylabel('P')
plt.subplot(325)
for i in range(10):
plt.plot(pars['range_t'], gE_bar_update[i, :])
plt.xlabel('Time (ms)')
plt.ylabel(r'$\bar g$')
plt.subplot(326)
plt.plot(pars['range_t'], gE, 'r')
plt.xlabel('Time (ms)')
plt.ylabel(r'$g_E$')
plt.tight_layout()
plt.show()
Think! 2A: Analyzing Synaptic Strength and Conductance Transformations#
In the above, even though all the presynaptic neurons have the same average firing rate, many of the synapses seem to have been weakened? Did you expect that?
Total synaptic conductance is fluctuating over time. How do you expect \(g_E\) to fluctuate if synapses did not show any STDP like behavior?
Do synaptic weights ever reach a stationary state when synapses show STDP?
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Analyzing_synaptic_strength_Discussion")
Distribution of synaptic weight#
From the example given above, we get an idea that some synapses depotentiate, but what is the distribution of the synaptic weights when synapses show STDP?
In fact, it is possible that even the synaptic weight distribution itself is a time-varying quantity. So, we would like to know how the distribution of synaptic weights evolves as a function of time.
To get a better estimate of the weight distribution and its time evolution, we will increase the presynaptic firing rate to \(15\)Hz and simulate the postsynaptic neuron for \(120\)s.
Functions for simulating a LIF neuron with STDP synapses#
Show code cell source
# @title Functions for simulating a LIF neuron with STDP synapses
def example_LIF_STDP(inputrate=15., Tsim=120000.):
"""
Simulation of a LIF model with STDP synapses
Args:
intputrate : The rate used for generate presynaptic spike trains
Tsim : Total simulation time
output:
Interactive demo, Visualization of synaptic weights
"""
pars = default_pars_STDP(T=Tsim, dt=1.)
pars['gE_bar'] = 0.024
pars['gE_init'] = 0.014 # initial synaptic conductance
pars['VE'] = 0. # [mV]
pars['tau_syn_E'] = 5. # [ms]
starttime = time.perf_counter()
pre_spike_train_ex = Poisson_generator(pars, rate=inputrate, n=300,
myseed=2020) # generate Poisson trains
v, rec_spikes, gE, P, M, gE_bar_update = run_LIF_cond_STDP(pars,
pre_spike_train_ex) # simulate LIF neuron with STDP
gbar_norm = gE_bar_update/pars['gE_bar'] # calculate the ratio of the synaptic conductance
endtime = time.perf_counter()
timecost = (endtime - starttime) / 60.
print('Total simulation time is %.3f min' % timecost)
my_layout.width = '620px'
@widgets.interact(
sample_time=widgets.FloatSlider(0.5, min=0., max=1., step=0.1,
layout=my_layout)
)
def my_visual_STDP_distribution(sample_time=0.0):
sample_time = int(sample_time * pars['range_t'].size) - 1
sample_time = sample_time * (sample_time > 0)
plt.figure(figsize=(8, 8))
ax1 = plt.subplot(211)
for i in range(50):
ax1.plot(pars['range_t'][::1000] / 1000., gE_bar_update[i, ::1000], lw=1., alpha=0.7)
ax1.axvline(1e-3 * pars['range_t'][sample_time], 0., 1., color='k', ls='--')
ax1.set_ylim(0, 0.025)
ax1.set_xlim(-2, 122)
ax1.set_xlabel('Time (s)')
ax1.set_ylabel(r'$\bar{g}$')
bins = np.arange(-.05, 1.05, .05)
g_dis, _ = np.histogram(gbar_norm[:, sample_time], bins)
ax2 = plt.subplot(212)
ax2.bar(bins[1:], g_dis, color='b', alpha=0.5, width=0.05)
ax2.set_xlim(-0.1, 1.1)
ax2.set_xlabel(r'$\bar{g}/g_{\mathrm{max}}$')
ax2.set_ylabel('Number')
ax2.set_title(('Time = %.1f s' % (1e-3 * pars['range_t'][sample_time])),
fontweight='bold')
plt.show()
print(help(example_LIF_STDP))
Help on function example_LIF_STDP in module __main__:
example_LIF_STDP(inputrate=15.0, Tsim=120000.0)
Simulation of a LIF model with STDP synapses
Args:
intputrate : The rate used for generate presynaptic spike trains
Tsim : Total simulation time
output:
Interactive demo, Visualization of synaptic weights
None
Interactive Demo 2: Example of an LIF model with STDP#
Make sure you execute this cell to enable the widget!
Show code cell source
# @markdown Make sure you execute this cell to enable the widget!
example_LIF_STDP(inputrate=15)
Think! 2B: Effects of Increased Presynaptic Firing Rate#
Increase the firing rate (i.e., 30 Hz) of presynaptic neurons, and investigate the effect on the dynamics of synaptic weight distribution.
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_LIF_and_STDP_Interactive_Demo_and_Discussion")
Section 3: Effect of input correlations#
Thus far, we assumed that the input population was uncorrelated. What do you think will happen if presynaptic neurons were correlated?
In the following, we will modify the input such that first \(L\) neurons have identical spike trains while the remaining inputs are uncorrelated. This is a highly simplified model of introducing correlations. You can try to code your own model of correlated spike trains.
Think! 3: Applications of STDP#
Modify the code above and create two groups of correlated presynaptic neurons and test what happens to the weight distribution.
How can the above observations be used to create unsupervised learning? Could you imagine how we have to train a neuronal model enabled with STDP rule to identify input patterns?
What else can be done with this type of plasticity?
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_LIF_plasticity_correlated_inputs_Interactive_Demo_and_Discussion")
Summary#
Hooray! You have just finished this loooong day! In this tutorial, we covered the concept of spike-timing dependent plasticity (STDP).
We managed to:
build a model of synapse that shows STDP.
study how correlations in input spike trains influence the distribution of synaptic weights.
Using presynaptic inputs as Poisson type spike trains, we modeled an LIF model with synapses equipped with STDP. We also studied the effect of correlated inputs on the synaptic strength!

