Tutorial 3: Building and Evaluating Normative Encoding Models#
Week 1, Day 5: Deep Learning
By Neuromatch Academy
Content creators: Jorge A. Menendez, Yalda Mohsenzadeh, Carsen Stringer
Content reviewers: Roozbeh Farhoodi, Madineh Sarvestani, Kshitij Dwivedi, Spiros Chavlis, Ella Batty, Michael Waskom
Production editors: Spiros Chavlis
Tutorial Objectives#
Estimated timing of tutorial: 1 hour, 10 minutes
In this tutorial, we’ll be using deep learning to build an encoding model of the visual system, and then compare its internal representations to those observed in neural data.
Its parameters won’t be directly optimized to fit the neural data. Instead, we will optimize its parameters to solve a particular visual task that we know the brain can solve. We therefore refer to it as a “normative” encoding model, since it is optimized for a specific behavioral task. It is the optimal model to solve the problem (optimal for the specified architecture). See Section 3 of the bonus tutorial to fit a convolutional neural network directly to neural data (an alternative approach for investigating encoding with deep neural networks).
To then evaluate whether this normative encoding model is actually a good model of the brain, we’ll analyze its internal representations and compare them to the representations observed in the mouse primary visual cortex. Since we understand exactly what the encoding model’s representations are optimized to do, any similarities will hopefully shed light on why the representations in the brain look the way they do.
More concretely, our goal will be learn how to:
Visualize and analyze the internal representations of a deep network
Quantify the similarity between distributed representations in a model and neural representations observed in recordings, using Representational Similarity Analysis (RSA)
Setup#
Install and import feedback gadget#
Show code cell source
# @title Install and import feedback gadget
!pip3 install vibecheck datatops --quiet
from vibecheck import DatatopsContentReviewContainer
def content_review(notebook_section: str):
return DatatopsContentReviewContainer(
"", # No text prompt
notebook_section,
{
"url": "https://pmyvdlilci.execute-api.us-east-1.amazonaws.com/klab",
"name": "neuromatch_cn",
"user_key": "y1x3mpx5",
},
).render()
feedback_prefix = "W1D5_T3"
# Imports
import numpy as np
from scipy.stats import zscore
import matplotlib as mpl
from matplotlib import pyplot as plt
import torch
from torch import nn, optim
from sklearn.decomposition import PCA
from sklearn.manifold import TSNE
Figure Settings#
Show code cell source
# @title Figure Settings
import logging
logging.getLogger('matplotlib.font_manager').disabled = True
%matplotlib inline
%config InlineBackend.figure_format='retina'
plt.style.use("https://raw.githubusercontent.com/NeuromatchAcademy/course-content/main/nma.mplstyle")
Plotting Functions#
Show code cell source
# @title Plotting Functions
def show_stimulus(img, ax=None, show=False):
"""Visualize a stimulus"""
if ax is None:
ax = plt.gca()
ax.imshow(img, cmap=mpl.cm.binary)
ax.set_aspect('auto')
ax.set_xticks([])
ax.set_yticks([])
ax.spines['left'].set_visible(False)
ax.spines['bottom'].set_visible(False)
if show:
plt.show()
def plot_corr_matrix(rdm, ax=None, show=False):
"""Plot dissimilarity matrix
Args:
rdm (numpy array): n_stimuli x n_stimuli representational dissimilarity
matrix
ax (matplotlib axes): axes onto which to plot
Returns:
nothing
"""
if ax is None:
ax = plt.gca()
image = ax.imshow(rdm, vmin=0.0, vmax=2.0)
ax.set_xticks([])
ax.set_yticks([])
cbar = plt.colorbar(image, ax=ax, label='dissimilarity')
if show:
plt.show()
def plot_multiple_rdm(rdm_dict):
"""Draw multiple subplots for each RDM in rdm_dict."""
fig, axs = plt.subplots(1, len(rdm_dict),
figsize=(4 * len(resp_dict), 3.5))
# Compute RDM's for each set of responses and plot
for i, (label, rdm) in enumerate(rdm_dict.items()):
image = plot_corr_matrix(rdm, axs[i])
axs[i].set_title(label)
plt.show()
def plot_rdm_rdm_correlations(rdm_sim):
"""Draw a bar plot showing between-RDM correlations."""
f, ax = plt.subplots()
ax.bar(rdm_sim.keys(), rdm_sim.values())
ax.set_xlabel('Deep network model layer')
ax.set_ylabel('Correlation of model layer RDM\nwith mouse V1 RDM')
plt.show()
def plot_rdm_rows(ori_list, rdm_dict, rdm_oris):
"""Plot the dissimilarity of response to each stimulus with response to one
specific stimulus
Args:
ori_list (list of float): plot dissimilarity with response to stimulus with
orientations closest to each value in this list
rdm_dict (dict): RDM's from which to extract dissimilarities
rdm_oris (np.ndarray): orientations corresponding to each row/column of RDMs
in rdm_dict
"""
n_col = len(ori_list)
f, axs = plt.subplots(1, n_col, figsize=(4 * n_col, 4), sharey=True)
# Get index of orientation closest to ori_plot
for ax, ori_plot in zip(axs, ori_list):
iori = np.argmin(np.abs(rdm_oris - ori_plot))
# Plot dissimilarity curves in each RDM
for label, rdm in rdm_dict.items():
ax.plot(rdm_oris, rdm[iori, :], label=label)
# Draw vertical line at stimulus we are plotting dissimilarity w.r.t.
ax.axvline(rdm_oris[iori], color=".7", zorder=-1)
# Label axes
ax.set_title(f'Dissimilarity with response\nto {ori_plot: .0f}$^o$ stimulus')
ax.set_xlabel('Stimulus orientation ($^o$)')
axs[0].set_ylabel('Dissimilarity')
axs[-1].legend(loc="upper left", bbox_to_anchor=(1, 1))
plt.tight_layout()
plt.show()
Helper Functions#
Show code cell source
# @title Helper Functions
def load_data(data_name, bin_width=1):
"""Load mouse V1 data from Stringer et al. (2019)
Data from study reported in this preprint:
https://www.biorxiv.org/content/10.1101/679324v2.abstract
These data comprise time-averaged responses of ~20,000 neurons
to ~4,000 stimulus gratings of different orientations, recorded
through Calcium imaginge. The responses have been normalized by
spontaneous levels of activity and then z-scored over stimuli, so
expect negative numbers. They have also been binned and averaged
to each degree of orientation.
This function returns the relevant data (neural responses and
stimulus orientations) in a torch.Tensor of data type torch.float32
in order to match the default data type for nn.Parameters in
Google Colab.
This function will actually average responses to stimuli with orientations
falling within bins specified by the bin_width argument. This helps
produce individual neural "responses" with smoother and more
interpretable tuning curves.
Args:
bin_width (float): size of stimulus bins over which to average neural
responses
Returns:
resp (torch.Tensor): n_stimuli x n_neurons matrix of neural responses,
each row contains the responses of each neuron to a given stimulus.
As mentioned above, neural "response" is actually an average over
responses to stimuli with similar angles falling within specified bins.
stimuli: (torch.Tensor): n_stimuli x 1 column vector with orientation
of each stimulus, in degrees. This is actually the mean orientation
of all stimuli in each bin.
"""
with np.load(data_name) as dobj:
data = dict(**dobj)
resp = data['resp']
stimuli = data['stimuli']
if bin_width > 1:
# Bin neural responses and stimuli
bins = np.digitize(stimuli, np.arange(0, 360 + bin_width, bin_width))
stimuli_binned = np.array([stimuli[bins == i].mean() for i in np.unique(bins)])
resp_binned = np.array([resp[bins == i, :].mean(0) for i in np.unique(bins)])
else:
resp_binned = resp
stimuli_binned = stimuli
# only use stimuli <= 180
resp_binned = resp_binned[stimuli_binned <= 180]
stimuli_binned = stimuli_binned[stimuli_binned <= 180]
stimuli_binned -= 90 # 0 means vertical, -ve means tilted left, +ve means tilted right
# Return as torch.Tensor
resp_tensor = torch.tensor(resp_binned, dtype=torch.float32)
stimuli_tensor = torch.tensor(stimuli_binned, dtype=torch.float32).unsqueeze(1) # add singleton dimension to make a column vector
return resp_tensor, stimuli_tensor
def grating(angle, sf=1 / 28, res=0.1, patch=False):
"""Generate oriented grating stimulus
Args:
angle (float): orientation of grating (angle from vertical), in degrees
sf (float): controls spatial frequency of the grating
res (float): resolution of image. Smaller values will make the image
smaller in terms of pixels. res=1.0 corresponds to 640 x 480 pixels.
patch (boolean): set to True to make the grating a localized
patch on the left side of the image. If False, then the
grating occupies the full image.
Returns:
torch.Tensor: (res * 480) x (res * 640) pixel oriented grating image
"""
angle = np.deg2rad(angle) # transform to radians
wpix, hpix = 640, 480 # width and height of image in pixels for res=1.0
xx, yy = np.meshgrid(sf * np.arange(0, wpix * res) / res, sf * np.arange(0, hpix * res) / res)
if patch:
gratings = np.cos(xx * np.cos(angle + .1) + yy * np.sin(angle + .1)) # phase shift to make it better fit within patch
gratings[gratings < 0] = 0
gratings[gratings > 0] = 1
xcent = gratings.shape[1] * .75
ycent = gratings.shape[0] / 2
xxc, yyc = np.meshgrid(np.arange(0, gratings.shape[1]), np.arange(0, gratings.shape[0]))
icirc = ((xxc - xcent) ** 2 + (yyc - ycent) ** 2) ** 0.5 < wpix / 3 / 2 * res
gratings[~icirc] = 0.5
else:
gratings = np.cos(xx * np.cos(angle) + yy * np.sin(angle))
gratings[gratings < 0] = 0
gratings[gratings > 0] = 1
# Return torch tensor
return torch.tensor(gratings, dtype=torch.float32)
def filters(out_channels=6, K=7):
""" make example filters, some center-surround and gabors
Returns:
filters: out_channels x K x K
"""
grid = np.linspace(-K/2, K/2, K).astype(np.float32)
xx,yy = np.meshgrid(grid, grid, indexing='ij')
# create center-surround filters
sigma = 1.1
gaussian = np.exp(-(xx**2 + yy**2)**0.5/(2*sigma**2))
wide_gaussian = np.exp(-(xx**2 + yy**2)**0.5/(2*(sigma*2)**2))
center_surround = gaussian - 0.5 * wide_gaussian
# create gabor filters
thetas = np.linspace(0, 180, out_channels-2+1)[:-1] * np.pi/180
gabors = np.zeros((len(thetas), K, K), np.float32)
lam = 10
phi = np.pi/2
gaussian = np.exp(-(xx**2 + yy**2)**0.5/(2*(sigma*0.4)**2))
for i,theta in enumerate(thetas):
x = xx*np.cos(theta) + yy*np.sin(theta)
gabors[i] = gaussian * np.cos(2*np.pi*x/lam + phi)
filters = np.concatenate((center_surround[np.newaxis,:,:],
-1*center_surround[np.newaxis,:,:],
gabors),
axis=0)
filters /= np.abs(filters).max(axis=(1,2))[:,np.newaxis,np.newaxis]
# convert to torch
filters = torch.from_numpy(filters)
# add channel axis
filters = filters.unsqueeze(1)
return filters
class CNN(nn.Module):
"""Deep convolutional network with one convolutional + pooling layer followed
by one fully connected layer
Args:
h_in (int): height of input image, in pixels (i.e. number of rows)
w_in (int): width of input image, in pixels (i.e. number of columns)
Attributes:
conv (nn.Conv2d): filter weights of convolutional layer
pool (nn.MaxPool2d): max pooling layer
dims (tuple of ints): dimensions of output from pool layer
fc (nn.Linear): weights and biases of fully connected layer
out (nn.Linear): weights and biases of output layer
"""
def __init__(self, h_in, w_in):
super().__init__()
C_in = 1 # input stimuli have only 1 input channel
C_out = 6 # number of output channels (i.e. of convolutional kernels to convolve the input with)
K = 7 # size of each convolutional kernel
Kpool = 8 # size of patches over which to pool
self.conv = nn.Conv2d(C_in, C_out, kernel_size=K, padding=K//2) # add padding to ensure that each channel has same dimensionality as input
self.pool = nn.MaxPool2d(Kpool)
self.dims = (C_out, h_in // Kpool, w_in // Kpool) # dimensions of pool layer output
self.fc = nn.Linear(np.prod(self.dims), 10) # flattened pool output --> 10D representation
self.out = nn.Linear(10, 1) # 10D representation --> scalar
self.conv.weight = nn.Parameter(filters(C_out, K))
self.conv.bias = nn.Parameter(torch.zeros((C_out,), dtype=torch.float32))
def forward(self, x):
"""Classify grating stimulus as tilted right or left
Args:
x (torch.Tensor): p x 48 x 64 tensor with pixel grayscale values for
each of p stimulus images.
Returns:
torch.Tensor: p x 1 tensor with network outputs for each input provided
in x. Each output should be interpreted as the probability of the
corresponding stimulus being tilted right.
"""
x = x.unsqueeze(1) # p x 1 x 48 x 64, add a singleton dimension for the single stimulus channel
x = torch.relu(self.conv(x)) # output of convolutional layer
x = self.pool(x) # output of pooling layer
x = x.view(-1, np.prod(self.dims)) # flatten pooling layer outputs into a vector
x = torch.relu(self.fc(x)) # output of fully connected layer
x = torch.sigmoid(self.out(x)) # network output
return x
def train(net, train_data, train_labels,
n_epochs=25, learning_rate=0.0005,
batch_size=100, momentum=.99):
"""Run stochastic gradient descent on binary cross-entropy loss for a given
deep network (cf. appendix for details)
Args:
net (nn.Module): deep network whose parameters to optimize with SGD
train_data (torch.Tensor): n_train x h x w tensor with stimulus gratings
train_labels (torch.Tensor): n_train x 1 tensor with true tilt of each
stimulus grating in train_data, i.e. 1. for right, 0. for left
n_epochs (int): number of times to run SGD through whole training data set
batch_size (int): number of training data samples in each mini-batch
learning_rate (float): learning rate to use for SGD updates
momentum (float): momentum parameter for SGD updates
"""
# Initialize binary cross-entropy loss function
loss_fn = nn.BCELoss()
# Initialize SGD optimizer with momentum
optimizer = optim.SGD(net.parameters(), lr=learning_rate, momentum=momentum)
# Placeholder to save loss at each iteration
track_loss = []
# Loop over epochs
for i in range(n_epochs):
# Split up training data into random non-overlapping mini-batches
ishuffle = torch.randperm(train_data.shape[0]) # random ordering of training data
minibatch_data = torch.split(train_data[ishuffle], batch_size) # split train_data into minibatches
minibatch_labels = torch.split(train_labels[ishuffle], batch_size) # split train_labels into minibatches
# Loop over mini-batches
for stimuli, tilt in zip(minibatch_data, minibatch_labels):
# Evaluate loss and update network weights
out = net(stimuli) # predicted probability of tilt right
loss = loss_fn(out, tilt) # evaluate loss
optimizer.zero_grad() # clear gradients
loss.backward() # compute gradients
optimizer.step() # update weights
# Keep track of loss at each iteration
track_loss.append(loss.item())
# Track progress
if (i + 1) % (n_epochs // 5) == 0:
print(f'epoch {i + 1} | loss on last mini-batch: {loss.item(): .2e}')
print('training done!')
def get_hidden_activity(net, stimuli, layer_labels):
"""Retrieve internal representations of network
Args:
net (nn.Module): deep network
stimuli (torch.Tensor): p x 48 x 64 tensor with stimuli for which to
compute and retrieve internal representations
layer_labels (list): list of strings with labels of each layer for which
to return its internal representations
Returns:
dict: internal representations at each layer of the network, in
numpy arrays. The keys of this dict are the strings in layer_labels.
"""
# Placeholder
hidden_activity = {}
# Attach 'hooks' to each layer of the network to store hidden
# representations in hidden_activity
def hook(module, input, output):
module_label = list(net._modules.keys())[np.argwhere([module == m for m in net._modules.values()])[0, 0]]
if module_label in layer_labels: # ignore output layer
hidden_activity[module_label] = output.view(stimuli.shape[0], -1).detach().numpy()
hooks = [layer.register_forward_hook(hook) for layer in net.children()]
# Run stimuli through the network
pred = net(stimuli)
# Remove the hooks
[h.remove() for h in hooks]
return hidden_activity
Data retrieval and loading#
Show code cell source
#@title Data retrieval and loading
import os
import hashlib
import requests
fname = "W3D4_stringer_oribinned1.npz"
url = "https://osf.io/683xc/download"
expected_md5 = "436599dfd8ebe6019f066c38aed20580"
if not os.path.isfile(fname):
try:
r = requests.get(url)
except requests.ConnectionError:
print("!!! Failed to download data !!!")
else:
if r.status_code != requests.codes.ok:
print("!!! Failed to download data !!!")
elif hashlib.md5(r.content).hexdigest() != expected_md5:
print("!!! Data download appears corrupted !!!")
else:
with open(fname, "wb") as fid:
fid.write(r.content)
Section 1: Setting up deep network and neural data#
In the future sections, we will compare the activity in a deep network, specifically in a CNN, with neural activity. First, we need to understand the task we are using (Section 1.1), train our deep network (Section 1.2), and load in neural data (Section 1.3).
Video 1: Deep convolutional network for orientation discrimination#
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Deep_convolutional_network_for_orientation_discrimination_Video")
Section 1.1: Orientation discrimination task#
We will build our normative encoding model by optimizing its parameters to solve an orientation discrimination task.
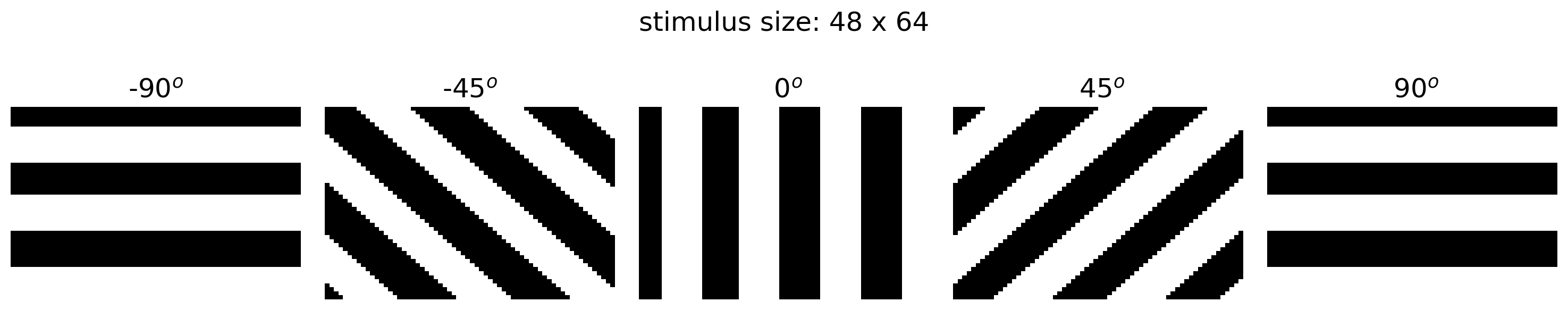
The task is to tell whether a given grating stimulus is tilted to the “right” or “left”; that is, whether its angle relative to the vertical is positive or negative, respectively. We show example stimuli below, which were constructed using the helper function grating().
Note that this is a task that we know many mammalian visual systems are capable of solving. It is therefore conceivable that the representations in a deep network model optimized for this task might resemble those in the brain. To test this hypothesis, we will compare the representations of our optimized encoding model to neural activity recorded in response to these very same stimuli, courtesy of Stringer et al 2019.
Execute this cell to plot example stimuli
Show code cell source
# @markdown Execute this cell to plot example stimuli
orientations = np.linspace(-90, 90, 5)
h_ = 3
n_col = len(orientations)
h, w = grating(0).shape # height and width of stimulus
fig, axs = plt.subplots(1, n_col, figsize=(h_ * n_col, h_))
for i, ori in enumerate(orientations):
stimulus = grating(ori)
axs[i].set_title(f'{ori: .0f}$^o$')
show_stimulus(stimulus, axs[i])
fig.suptitle(f'stimulus size: {h} x {w}')
plt.tight_layout()
plt.show()
Section 1.2: A deep network model of orientation discrimination#
Estimated timing to here from start of tutorial: 10 min
Our goal is to build a model that solves the orientation discrimination task outlined above. The model should take as input a stimulus image and output the probability of that stimulus being tilted right.
To do this, we will use a convolutional neural network (CNN), which is the type of network we saw in Tutorial 2. Here, we will use a CNN that performs two-dimensional convolutions on the raw stimulus image (which is a 2D matrix of pixels), rather than one-dimensional convolutions on a categorical 1D vector representation of the stimulus. CNNs are commonly used for image processing.
The particular CNN we will use here has two layers:
a convolutional layer, which convolves the images with a set of filters
a fully connected layer, which transforms the output of this convolution into a 10-dimensional representation
Finally, a set of output weights transforms this 10-dimensional representation into a single scalar \(p\), denoting the predicted probability of the input stimulus being tilted right.
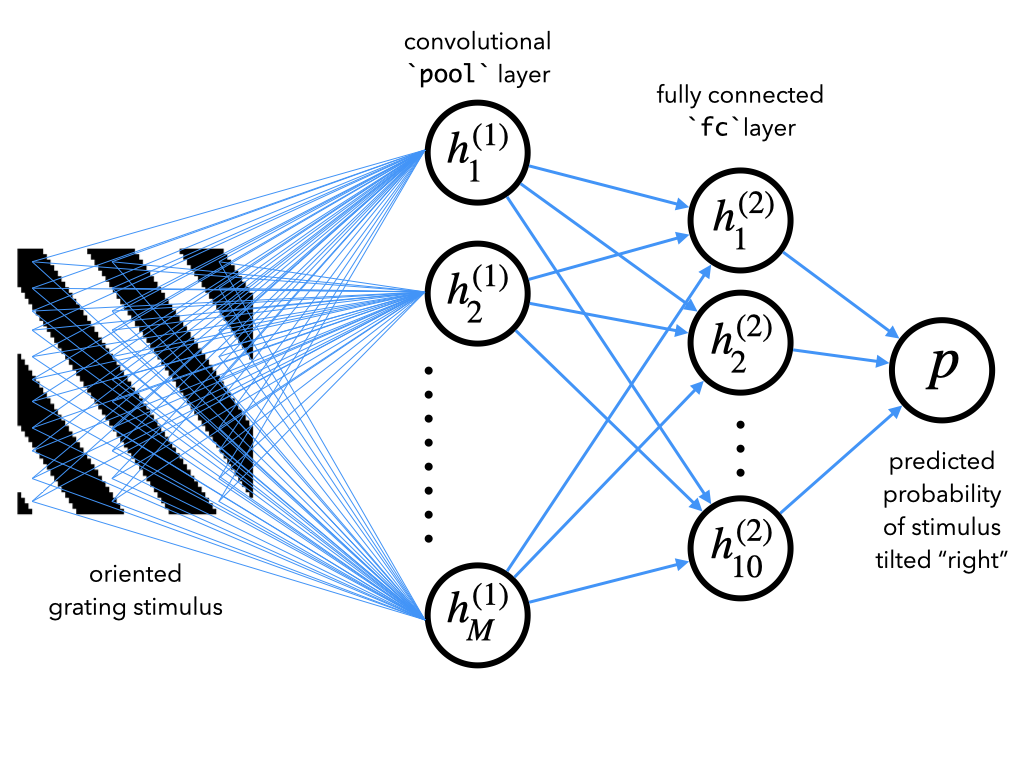
See bonus section 1 for in-depth instructions for how to code up such a network in PyTorch. For now, however, we’ll leave these details aside and focus on training this network, called CNN, and analyzing its internal representations.
Run the next cell to train such a network to solve this task. After initializing our CNN model, it builds a dataset of oriented grating stimuli to use for training it. These are then passed into a function called train() that uses SGD to optimize the model’s parameters, taking similar arguments as the train() function we wrote in Tutorial 1.
Note that it may take ~30 seconds for the training to complete.
help(train)
Help on function train in module __main__:
train(net, train_data, train_labels, n_epochs=25, learning_rate=0.0005, batch_size=100, momentum=0.99)
Run stochastic gradient descent on binary cross-entropy loss for a given
deep network (cf. appendix for details)
Args:
net (nn.Module): deep network whose parameters to optimize with SGD
train_data (torch.Tensor): n_train x h x w tensor with stimulus gratings
train_labels (torch.Tensor): n_train x 1 tensor with true tilt of each
stimulus grating in train_data, i.e. 1. for right, 0. for left
n_epochs (int): number of times to run SGD through whole training data set
batch_size (int): number of training data samples in each mini-batch
learning_rate (float): learning rate to use for SGD updates
momentum (float): momentum parameter for SGD updates
# Set random seeds for reproducibility
np.random.seed(12)
torch.manual_seed(12)
# Initialize CNN model
net = CNN(h, w)
# Build training set to train it on
n_train = 1000 # size of training set
# sample n_train random orientations between -90 and +90 degrees
ori = (np.random.rand(n_train) - 0.5) * 180
# build orientated grating stimuli
stimuli = torch.stack([grating(i) for i in ori])
# stimulus tilt: 1. if tilted right, 0. if tilted left, as a column vector
tilt = torch.tensor(ori > 0).type(torch.float).unsqueeze(-1)
# Train model
train(net, stimuli, tilt)
epoch 5 | loss on last mini-batch: 4.10e-01
epoch 10 | loss on last mini-batch: 5.17e-02
epoch 15 | loss on last mini-batch: 1.32e-02
epoch 20 | loss on last mini-batch: 2.29e-03
epoch 25 | loss on last mini-batch: 8.08e-05
training done!
Section 1.3: Load data#
Estimated timing to here from start of tutorial: 15 min
In the next cell, we provide code for loading in some data from Stringer et al., 2021, which contains the responses of about ~20,000 neurons in the mouse primary visual cortex to grating stimuli like those used to train our network (this is the same data used in Tutorial 1). These data are stored in two variables:
resp_v1is a matrix where each row contains the responses of all neurons to a single stimulus.oriis a vector with the orientations of each stimulus, in degrees. As in the above convention, negative angles denote stimuli tilted to the left and positive angles denote stimuli tilted to the right.
We will then extract our deep CNN model’s representations of these same stimuli (i.e. oriented gratings with the orientations in ori). We will run the same stimuli through our CNN model and use the helper function get_hidden_activity() to store the model’s internal representations. The output of this function is a Python dict, which contains a matrix of population responses (just like resp_v1) for each layer of the network specified by the layer_labels argument. We’ll focus on looking at the representations in
the output of the first convolutional layer, stored in the model as
'pool'(see Bonus Section 1 for the details of the CNN architecture to understand why it’s called this way)the 10-dimensional output of the fully connected layer, stored in the model as
'fc'
# Load mouse V1 data
resp_v1, ori = load_data(fname)
# Extract model internal representations of each stimulus in the V1 data
# construct grating stimuli for each orientation presented in the V1 data
stimuli = torch.stack([grating(a.item()) for a in ori])
layer_labels = ['pool', 'fc']
resp_model = get_hidden_activity(net, stimuli, layer_labels)
# Aggregate all responses into one dict
resp_dict = {}
resp_dict['V1 data'] = resp_v1
for k, v in resp_model.items():
label = f"model\n'{k}' layer"
resp_dict[label] = v
Section 2: Quantitative comparisons of CNNs and neural activity#
Estimated timing to here from start of tutorial: 20 min
Let’s now analyze the internal representations of our deep CNN model of orientation discrimination and compare them to population responses in the mouse primary visual cortex.
In this section, we’ll try to quantitatively compare CNN and primary visual cortex representations. In the next section, we will visualize their representations and get some intuition for their structure.
Video 2: Quantitative comparisons of CNNs and neural activity#
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Quantitative_comparisons_of_CNNs_and_neural_activity_Video")
We noticed above some similarities and differences between the population responses in the mouse primary visual cortex and in different layers in our model. Let’s now try to quantify this.
To do this, we’ll use a technique called Representational Similarity Analysis. The idea is to look at the similarity structure between representations of different stimuli. We can say that a brain area and a model use a similar representational scheme if stimuli that are represented (dis)similarly in the brain are represented (dis)similarly in the model as well.
Section 2.1: Representational dissimilarity matrix (RDM)#
To quantify this, we begin by computing the representational dissimilarity matrix (RDM) for the mouse V1 data and each model layer. This matrix, which we’ll call \(\mathbf{M}\), is computed as one minus the correlation coefficients between population responses to each stimulus. We can efficiently compute this by using the \(z\)-scored responses.
The \(z\)-scored response of all neurons \(\mathbf{r}\) to stimulus \(s\) is the response mean-subtracted across neurons \(i\) and normalized to standard deviation 1 across neurons \(i\) where \(N\) is the total number of neurons:
where \(\mu^{(s)} = \frac{1}{N}\sum_{i=1}^N r_i^{(s)}\) and \(\sigma^{(s)} = \sqrt{\frac{1}{N}\sum_{i=1}^N \left( r_i^{(s)} - \mu^{(s)} \right)^2}\).
Then the full matrix can be computed as:
where \(\mathbf{Z}\) is the z-scored response matrix with rows \(\mathbf{r}^{(s)}\) and N is the number of neurons (or units). See bonus section 3 for full explanation.
Coding Exercise 2.1: Compute RDMs#
Complete the function RDM() for computing the RDM for a given set of population responses to each stimulus. Use the above formula in terms of \(z\)-scored population responses. We will use the helper function zscore() to compute the matrix of \(z\)-scored responses.
The subsequent cell uses this function to plot the RDM of the population responses in the V1 data and in each layer of our model CNN.
def RDM(resp):
"""Compute the representational dissimilarity matrix (RDM)
Args:
resp (ndarray): S x N matrix with population responses to
each stimulus in each row
Returns:
ndarray: S x S representational dissimilarity matrix
"""
#########################################################
## TO DO for students: compute representational dissimilarity matrix
# Fill out function and remove
raise NotImplementedError("Student exercise: complete function RDM")
#########################################################
# z-score responses to each stimulus
zresp = zscore(resp, axis=1)
# Compute RDM
RDM = ...
return RDM
# Compute RDMs for each layer
rdm_dict = {label: RDM(resp) for label, resp in resp_dict.items()}
# Plot RDMs
plot_multiple_rdm(rdm_dict)
Example output:
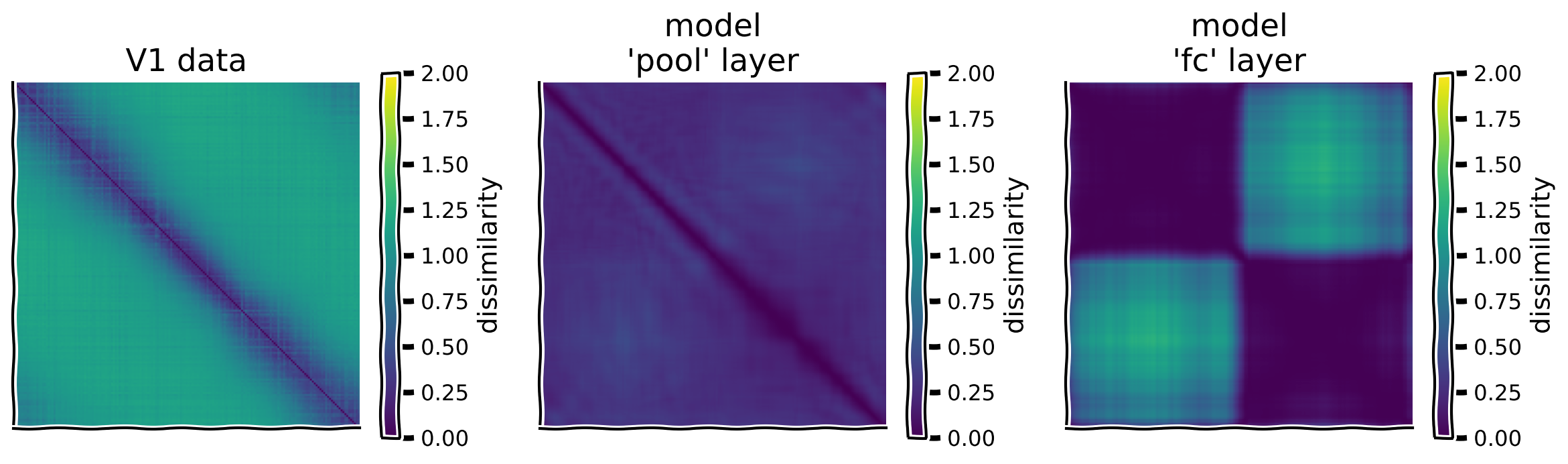
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Compute_RDMs_Exercise")
Video 3: Coding Exercise 2.1 solution discussion#
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Solution_Discussion_Video")
Section 2.2: Determining representation similarity#
Estimated timing to here from start of tutorial: 35 min
To quantify how similar the representations are, we can simply correlate their dissimilarity matrices. For this, we’ll again use the correlation coefficient. Note that dissimilarity matrices are symmetric (\(M_{ss'} = M_{s's}\)), so we should only use the off-diagonal terms on one side of the diagonal when computing this correlation to avoid overcounting. Moreover, we should leave out the diagonal terms, which are always equal to 0, so will always be perfectly correlated across any pair of RDM’s.
Coding Exercise 2.2: Correlate RDMs#
Complete the function correlate_rdms() below that computes this correlation. The code for extracting the off-diagonal terms is provided.
We will then use a function to compute the correlation between the RDM’s for each layer of our model CNN and that of the V1 data.
def correlate_rdms(rdm1, rdm2):
"""Correlate off-diagonal elements of two RDM's
Args:
rdm1 (np.ndarray): S x S representational dissimilarity matrix
rdm2 (np.ndarray): S x S representational dissimilarity matrix to
correlate with rdm1
Returns:
float: correlation coefficient between the off-diagonal elements
of rdm1 and rdm2
"""
# Extract off-diagonal elements of each RDM
ioffdiag = np.triu_indices(rdm1.shape[0], k=1) # indices of off-diagonal elements
rdm1_offdiag = rdm1[ioffdiag]
rdm2_offdiag = rdm2[ioffdiag]
#########################################################
## TO DO for students: compute correlation coefficient
# Fill out function and remove
raise NotImplementedError("Student exercise: complete correlate rdms")
#########################################################
corr_coef = np.corrcoef(..., ...)[0,1]
return corr_coef
# Split RDMs into V1 responses and model responses
rdm_model = rdm_dict.copy()
rdm_v1 = rdm_model.pop('V1 data')
# Correlate off-diagonal terms of dissimilarity matrices
rdm_sim = {label: correlate_rdms(rdm_v1, rdm) for label, rdm in rdm_model.items()}
# Visualize
plot_rdm_rdm_correlations(rdm_sim)
Example output:
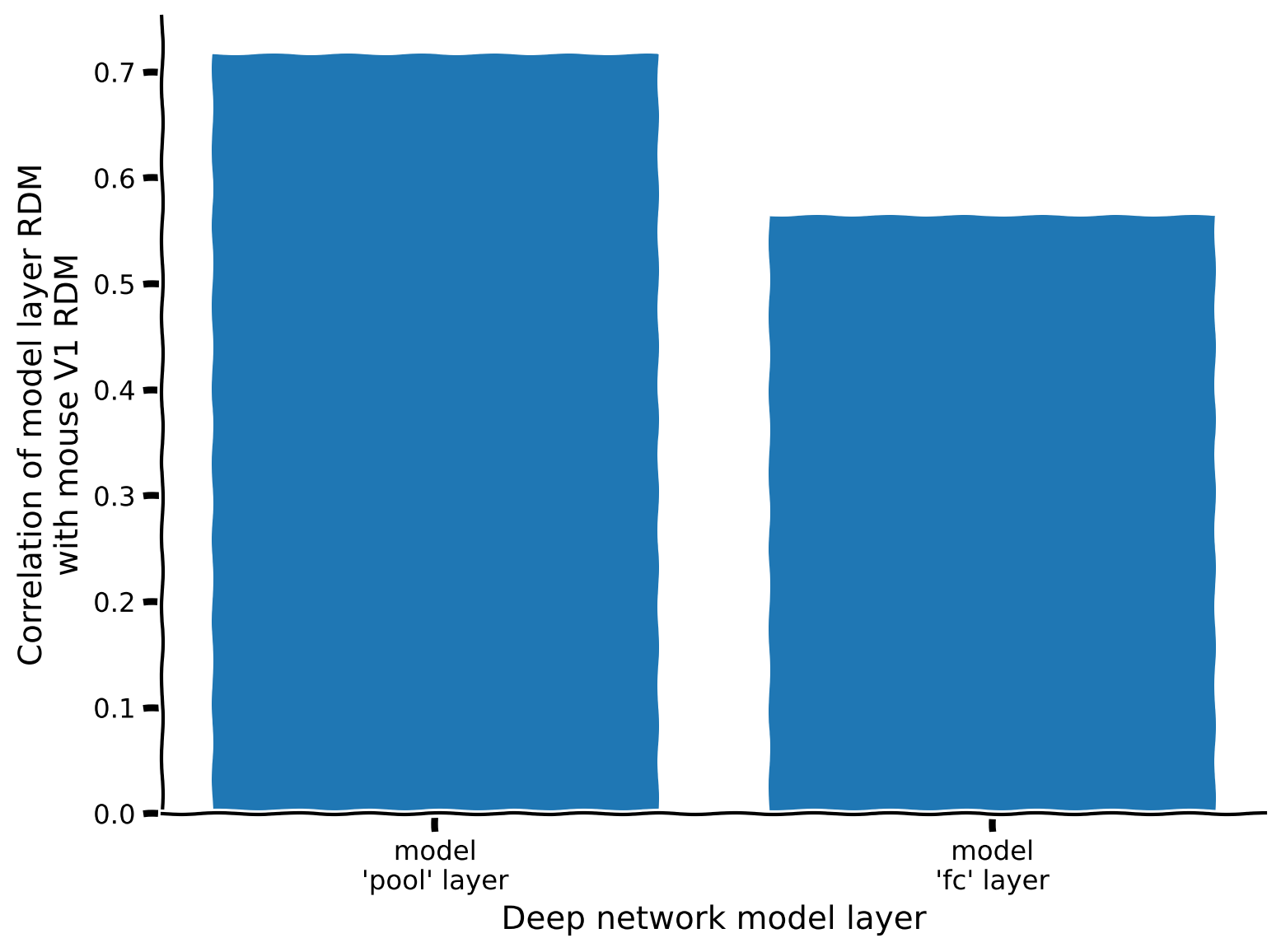
According to this metric, which layer’s representations most resemble those in the data? Does this agree with your intuitions from coding exercise 2.1?
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Correlate_RDMs_Exercise")
Section 2.3: Further understanding RDMs#
Estimated timing to here from start of tutorial: 45 min
To better understand how these correlations in RDM’s arise, we can try plotting individual rows of the RDM matrix. The resulting curves show the similarity of the responses to each stimulus with that to one specific stimulus.
ori_list = [-75, -25, 25, 75]
plot_rdm_rows(ori_list, rdm_dict, ori.numpy())
Section 3: Qualitative comparisons of CNNs and neural activity#
To visualize the representations in the data and in each of these model layers, we’ll use two classic techniques from systems neuroscience:
tuning curves: plotting the response of single neurons (or units, in the case of the deep network) as a function of the stimulus orientation
dimensionality reduction: plotting full population responses to each stimulus in two dimensions via dimensionality reduction. We’ll use the non-linear dimensionality reduction technique t-SNE for this. We use dimensionality reduction because there are many units and it’s difficult to visualize all of them at once. We use a non-linear dimensionality reduction technique because it can capture complex relationships between stimuli (see W1D5 for more details).
Section 3.1: Tuning Curves#
Estimated timing to here from start of tutorial: 50 min
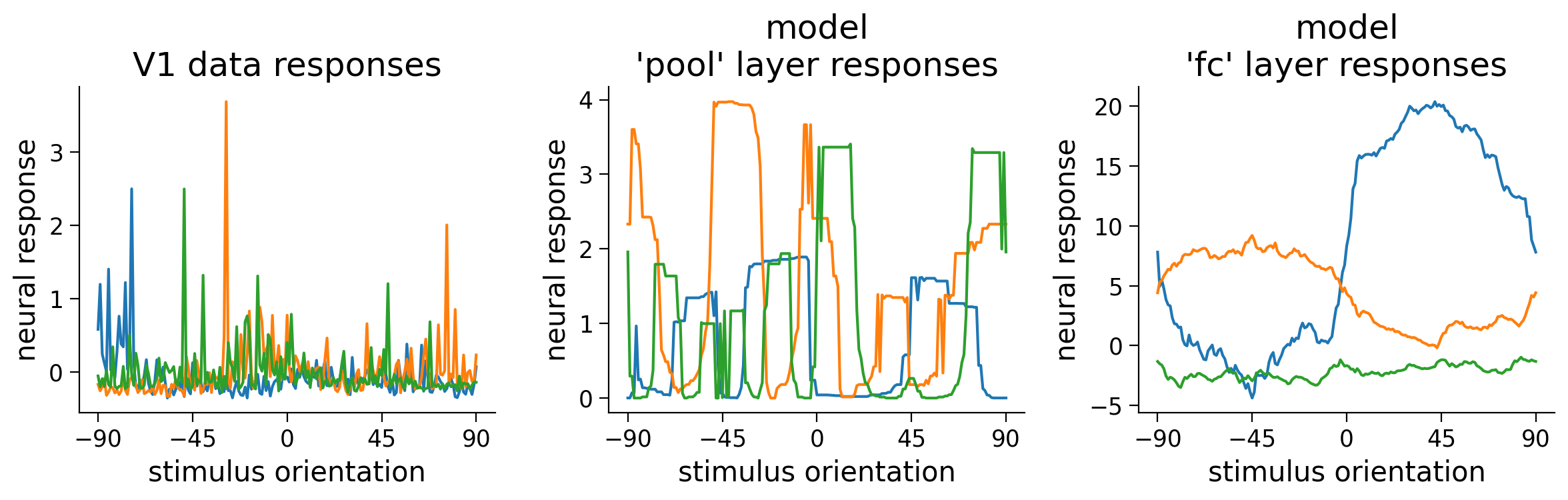
Below, we show some example tuning curves for different neurons and units in the CNN we trained above. How are the single neuron responses similar/different between the model and the data? Try running this cell multiple times to get an idea of shared properties in the tuning curves of the neurons within each population.
Execute this cell to visualize tuning curves
Show code cell source
# @markdown Execute this cell to visualize tuning curves
fig, axs = plt.subplots(1, len(resp_dict), figsize=(len(resp_dict) * 4, 4))
for i, (label, resp) in enumerate(resp_dict.items()):
ax = axs[i]
ax.set_title(f'{label} responses')
# Pick three random neurons whose tuning curves to plot
ineurons = np.random.choice(resp.shape[1], 3, replace=False)
# Plot tuning curves of ineurons
ax.plot(ori, resp[:, ineurons])
ax.set_xticks(np.linspace(-90, 90, 5))
ax.set_xlabel('stimulus orientation')
ax.set_ylabel('neural response')
plt.tight_layout()
plt.show()
Section 3.2: Dimensionality reduction of representations#
Estimated timing to here from start of tutorial: 55 min
We can visualize a dimensionality-reduced version of the internal representations of the mouse primary visual cortex or CNN internal representations in order to potentially uncover informative structure. Here, we use PCA to reduce the dimensionality to 20 dimensions, and then use tSNE to further reduce dimensionality to 2 dimensions. We use the first step of PCA so that tSNE runs faster (this is standard practice in the field).
Execute this cell to visualize low-d representations
Show code cell source
# @markdown Execute this cell to visualize low-d representations
def plot_resp_lowd(resp_dict):
"""Plot a low-dimensional representation of each dataset in resp_dict."""
n_col = len(resp_dict)
fig, axs = plt.subplots(1, n_col, figsize=(4.5 * len(resp_dict), 4.5))
for i, (label, resp) in enumerate(resp_dict.items()):
ax = axs[i]
ax.set_title(f'{label} responses')
# First do PCA to reduce dimensionality to 20 dimensions so that tSNE is faster
resp_lowd = PCA(n_components=min(20, resp.shape[1]), random_state=0).fit_transform(resp)
# Then do tSNE to reduce dimensionality to 2 dimensions
resp_lowd = TSNE(n_components=2, random_state=0).fit_transform(resp_lowd)
# Plot dimensionality-reduced population responses 'resp_lowd'
# on 2D axes, with each point colored by stimulus orientation
x, y = resp_lowd[:, 0], resp_lowd[:, 1]
pts = ax.scatter(x, y, c=ori, cmap='twilight', vmin=-90, vmax=90)
fig.colorbar(pts, ax=ax, ticks=np.linspace(-90, 90, 5),
label='Stimulus orientation')
ax.set_xlabel('Dimension 1')
ax.set_ylabel('Dimension 2')
ax.set_xticks([])
ax.set_yticks([])
plt.show()
plot_resp_lowd(resp_dict)
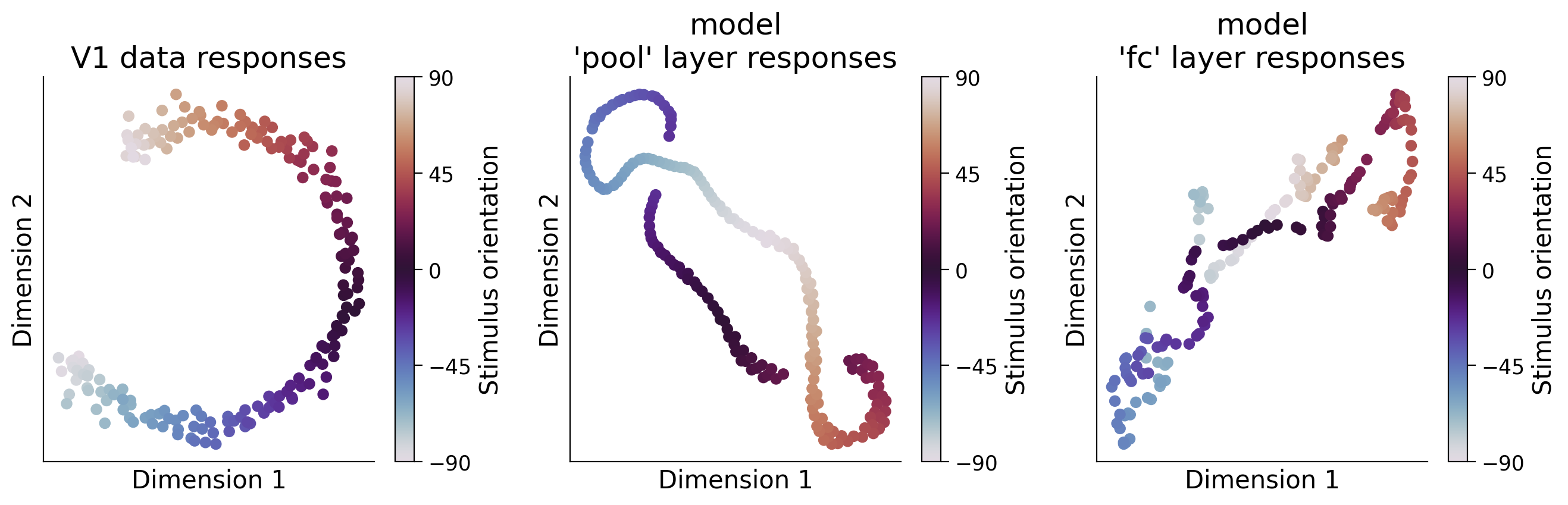
Think! 3.2: Visualizing reduced dimensionality representations#
Interpret the figure above. Why do these representations look the way they do? Here are a few specific questions to think about:
How are the population responses similar/different between the model and the data? Can you explain these population-level responses from the single neuron responses seen in the previous exercise, or vice-versa?
How do the representations in the different layers of the model differ, and how does this relate to the orientation discrimination task the model was optimized for?
Which layer of our deep network encoding model most closely resembles the V1 data?
Submit your feedback#
Show code cell source
# @title Submit your feedback
content_review(f"{feedback_prefix}_Vizualizing_reduced_dimensionality_representations_Discussion")
Summary#
Estimated timing of tutorial: 1 hour, 10 minutes
In this notebook, we learned
how to use deep learning to build a normative encoding model of the visual system
how to use RSA to evaluate how the model’s representations match to those in the brain
Our approach was to optimize a deep convolutional network to solve an orientation discrimination task. But note that many other approaches could have been taken.
Firstly, there are many other “normative” ways to solve this orientation discrimination task. We could have used different neural network architectures, or even used a completely different algorithm that didn’t involve a neural network at all, but instead used other kinds of image transformations (e.g. Fourier transforms). Neural network approaches, however, are special in that they explicitly use abstract distributed representations to compute, which feels a lot closer to the kinds of algorithms the brain uses. Convolutional neural networks in particular are well-suited for building normative models of the visual system.
Secondly, our choice of visual task was mostly arbitrary. For example, we could have trained our network to directly estimate the orientation of the stimulus, rather than just discriminating between two classes of tilt. Or, we could have trained the network to perform a more naturalistic task, such as recognizing the rotation of an arbitrary image. Or we could try a task like object recognition. Is this something that mice compute in their visual cortex?
Training on different tasks could lead to different representations of the oriented grating stimuli, which might match the observed V1 representations better or worse.
See Section 3 in the Bonus Tutorial to fit a convolutional neural network directly to neural activity to build an encoding model.
Bonus#
Bonus Section 1: Building CNN’s with PyTorch#
Here we walk through building the different types of layers in a CNN using PyTorch, culminating in the CNN model used above.
Bonus Section 1.1: Fully connected layers#
In a fully connected layer, each unit computes a weighted sum over all the input units and applies a non-linear function to this weighted sum. You have used such layers many times already in parts 1 and 2. As you have already seen, these are implemented in PyTorch using the nn.Linear class.
See the next cell for code for constructing a deep network with one fully connected layer that will classify an input image as being tilted left or right. Specifically, its output is the predicted probability of the input image being tilted right. To ensure that its output is a probability (i.e. a number between 0 and 1), we use a sigmoid activation function to squash the output into this range (implemented with torch.sigmoid()).
class FC(nn.Module):
"""Deep network with one fully connected layer
Args:
h_in (int): height of input image, in pixels (i.e. number of rows)
w_in (int): width of input image, in pixels (i.e. number of columns)
Attributes:
fc (nn.Linear): weights and biases of fully connected layer
out (nn.Linear): weights and biases of output layer
"""
def __init__(self, h_in, w_in):
super().__init__()
self.dims = h_in * w_in # dimensions of flattened input
self.fc = nn.Linear(self.dims, 10) # flattened input image --> 10D representation
self.out = nn.Linear(10, 1) # 10D representation --> scalar
def forward(self, x):
"""Classify grating stimulus as tilted right or left
Args:
x (torch.Tensor): p x 48 x 64 tensor with pixel grayscale values for
each of p stimulus images.
Returns:
torch.Tensor: p x 1 tensor with network outputs for each input provided
in x. Each output should be interpreted as the probability of the
corresponding stimulus being tilted right.
"""
x = x.view(-1, self.dims) # flatten each input image into a vector
x = torch.relu(self.fc(x)) # output of fully connected layer
x = torch.sigmoid(self.out(x)) # network output
return x
Bonus Section 1.2: Convolutional layers#
In a convolutional layer, each unit computes a weighted sum over a two-dimensional \(K \times K\) patch of inputs. As we saw in part 2, the units are arranged in channels (see figure below), whereby units in the same channel compute the same weighted sum over different parts of the input, using the weights of that channel’s convolutional filter (or kernel). The output of a convolutional layer is thus a three-dimensional tensor of shape \(C^{out} \times H \times W\), where \(C^{out}\) is the number of channels (i.e. the number of convolutional filters/kernels), and \(H\) and \(W\) are the height and width of the input.

Such layers can be implemented in Python using the PyTorch class `nn.Conv2d as we saw in Tutorial 2 (documentation here).
See the next cell for code incorporating a convolutional layer with 8 convolutional filters of size 5 \(\times\) 5 into our above fully connected network. Note that we have to flatten the multi-channel output in order to pass it on to the fully connected layer.
class ConvFC(nn.Module):
"""Deep network with one convolutional layer and one fully connected layer
Args:
h_in (int): height of input image, in pixels (i.e. number of rows)
w_in (int): width of input image, in pixels (i.e. number of columns)
Attributes:
conv (nn.Conv2d): filter weights of convolutional layer
dims (tuple of ints): dimensions of output from conv layer
fc (nn.Linear): weights and biases of fully connected layer
out (nn.Linear): weights and biases of output layer
"""
def __init__(self, h_in, w_in):
super().__init__()
C_in = 1 # input stimuli have only 1 input channel
C_out = 6 # number of output channels (i.e. of convolutional kernels to convolve the input with)
K = 7 # size of each convolutional kernel (should be odd number for the padding to work as expected)
self.conv = nn.Conv2d(C_in, C_out, kernel_size=K, padding=K//2) # add padding to ensure that each channel has same dimensionality as input
self.dims = (C_out, h_in, w_in) # dimensions of conv layer output
self.fc = nn.Linear(np.prod(self.dims), 10) # flattened conv output --> 10D representation
self.out = nn.Linear(10, 1) # 10D representation --> scalar
def forward(self, x):
"""Classify grating stimulus as tilted right or left
Args:
x (torch.Tensor): p x 48 x 64 tensor with pixel grayscale values for
each of p stimulus images.
Returns:
torch.Tensor: p x 1 tensor with network outputs for each input provided
in x. Each output should be interpreted as the probability of the
corresponding stimulus being tilted right.
"""
x = x.unsqueeze(1) # p x 1 x 48 x 64, add a singleton dimension for the single stimulus channel
x = torch.relu(self.conv(x)) # output of convolutional layer
x = x.view(-1, np.prod(self.dims)) # flatten convolutional layer outputs into a vector
x = torch.relu(self.fc(x)) # output of fully connected layer
x = torch.sigmoid(self.out(x)) # network output
return x
Bonus Section 1.3: Max pooling layers#
In a max pooling layer, each unit computes the maximum over a small two-dimensional \(K^{pool} \times K^{pool}\) patch of inputs. Given a multi-channel input of dimensions \(C \times H \times W\), the output of a max pooling layer has dimensions \(C \times H^{out} \times W^{out}\), where:
where \(\lfloor\cdot\rfloor\) denotes rounding down to the nearest integer below (i.e., floor division // in Python).
Max pooling layers can be implemented with the PyTorch nn.MaxPool2d class, which takes as a single argument the size \(K^{pool}\) of the pooling patch. See the next cell for an example, which builds upon the previous example by adding in a max pooling layer just after the convolutional layer. Note again that we need to calculate the dimensions of its output in order to set the dimensions of the subsequent fully connected layer.
class ConvPoolFC(nn.Module):
"""Deep network with one convolutional layer followed by a max pooling layer
and one fully connected layer
Args:
h_in (int): height of input image, in pixels (i.e. number of rows)
w_in (int): width of input image, in pixels (i.e. number of columns)
Attributes:
conv (nn.Conv2d): filter weights of convolutional layer
pool (nn.MaxPool2d): max pooling layer
dims (tuple of ints): dimensions of output from pool layer
fc (nn.Linear): weights and biases of fully connected layer
out (nn.Linear): weights and biases of output layer
"""
def __init__(self, h_in, w_in):
super().__init__()
C_in = 1 # input stimuli have only 1 input channel
C_out = 6 # number of output channels (i.e. of convolutional kernels to convolve the input with)
K = 7 # size of each convolutional kernel
Kpool = 8 # size of patches over which to pool
self.conv = nn.Conv2d(C_in, C_out, kernel_size=K, padding=K//2) # add padding to ensure that each channel has same dimensionality as input
self.pool = nn.MaxPool2d(Kpool)
self.dims = (C_out, h_in // Kpool, w_in // Kpool) # dimensions of pool layer output
self.fc = nn.Linear(np.prod(self.dims), 10) # flattened pool output --> 10D representation
self.out = nn.Linear(10, 1) # 10D representation --> scalar
def forward(self, x):
"""Classify grating stimulus as tilted right or left
Args:
x (torch.Tensor): p x 48 x 64 tensor with pixel grayscale values for
each of p stimulus images.
Returns:
torch.Tensor: p x 1 tensor with network outputs for each input provided
in x. Each output should be interpreted as the probability of the
corresponding stimulus being tilted right.
"""
x = x.unsqueeze(1) # p x 1 x 48 x 64, add a singleton dimension for the single stimulus channel
x = torch.relu(self.conv(x)) # output of convolutional layer
x = self.pool(x) # output of pooling layer
x = x.view(-1, np.prod(self.dims)) # flatten pooling layer outputs into a vector
x = torch.relu(self.fc(x)) # output of fully connected layer
x = torch.sigmoid(self.out(x)) # network output
return x
This pooling layer completes the CNN model trained above to perform orientation discrimination. We can think of this architecture as having two primary layers:
a convolutional + pooling layer
a fully connected layer
We group together the convolution and pooling layers into one, as they really form one full unit of convolutional processing, where each patch of the image is passed through a convolutional filter and pooled with neighboring patches. It is standard practice to follow up any convolutional layer with a pooling layer, so they are generally treated as a single block of processing.
Bonus Section 2: Orientation discrimination as a binary classification problem#
What loss function should we minimize to optimize orientation discrimination performance? We first note that the orientation discrimination task is a binary classification problem, where the goal is to classify a given stimulus into one of two classes: being tilted left or being tilted right.
Our goal is thus to output a high probability of the stimulus being tilted right (i.e. large \(p\)) whenever the stimulus is tilted right, and a high probability of the stimulus being tilted left (i.e. large \(1-p \Leftrightarrow\) small \(p\)) whenever the stimulus is tilted left.
Let \(\tilde{y}^{(n)}\) be the label of the \(n\)th stimulus in the mini-batch, indicating its true tilt:
Let \(p^{(n)}\) be the predicted probability of that stimulus being tilted right assigned by our network. Note that that \(1-p^{(n)}\) is the predicted probability of that stimulus being tilted left. We’d now like to modify the parameters so as to maximize the predicted probability of the true class \(\tilde{y}^{(n)}\). One way to formalize this is as maximizing the log probability
You should recognize this expression as the log likelihood of the Bernoulli distribution under the predicted probability \(p^{(n)}\). This is the same quantity that is maximized in logistic regression, where the predicted probability \(p^{(n)}\) is just a simple linear sum of its inputs (rather than a complicated non-linear operation, like in the deep networks used here).
To turn this into a loss function, we simply multiply it by -1, resulting in the so-called binary cross-entropy, or negative log likelihood. Summing over \(P\) samples in a batch, the binary cross entropy loss is given by
The binary cross-entropy loss can be implemented in PyTorch using the nn.BCELoss() loss function (cf. documentation).
Feel free to check out the code used to optimize the CNN in the train() function defined in the hidden cell of helper functions at the top of the notebook. Because the CNN’s used here have lots of parameters, we have to use two tricks that we didn’t use in the previous parts of this tutorial:
We have to use stochastic gradient descent (SGD), rather than just gradient descent (GD).
We have to use momentum in our SGD updates. This is easily incorporated into our PyTorch implementation by just setting the
momentumargument of the built-inoptim.SGDoptimizer.
Bonus Section 3: RDM Z-Score Explanation#
If \(r^{(s)}_i\) is the response of the \(i\)-th neuron to the \(s\)-th stimulus, then
This can be computed efficiently by using the \(z\)-scored responses
such that the full matrix can be computed through the matrix multiplication
where \(S\) is the total number of stimuli. Note that \(\mathbf{Z}\) is an \(S \times N\) matrix, and \(\mathbf{M}\) is an \(S \times S\) matrix.

